BactoBox® Application Note
Why You Should Choose BactoBox® over OD600
The plate count method is by far the most typical method for determining the number of viable bacteria in a sample and therefore often stated as the gold standard method for this purpose |1|. Unfortunately - even for some of the fastest growing bacteria - it typically takes more than 10 hours for colonies to appear to the naked eye. Plate counts do therefore not provide actionable here-and-now knowledge for fermentation processes. Optical density at 600 nm (OD600) is one of the most frequently used proxy methods for plate counts, but it is well-known that there are pitfalls with this technique. BactoBox represents a novel rapid microbiology alternative to track in-process fermentation samples and bacterial test suspensions. The data provided here for an E. coli growth curve show that BactoBox detects growth sooner than OD600 and clearly provides more consistent correlations with plate counts.
Keywords: Spectrophotometry, spectrophotometer, OD600, OD660, dry weight estimates, biomass estimates, particles, light scattering, impedance flow cytometry, impedance spectroscopy, rapid microbiology methods, cell counter, cell counts, total viable count, turbidity, nephelometry, wavelength, absorbance, biomass monitoring, microbial fermentation, scientific bioprocessing, microbial upstream bioprocessing, McFarland standards.
It is always a pleasure to engage with customers and satisfy your curiosity. We are often asked very interesting questions, and this list is an attempt to answer some of the most common ones. However, there are always multiple layers to a certain topic and question, so do not hesitate to follow up if you desire a more in-depth explanation to any of the topics below!
Is the measurement real-time or how fast is it?
When a bacterium passes over the electrodes in our flow cell it will be detected. This detection happens in real-time. However, we only measure a subset of the sample inserted in BactoBox. We need to measure for a certain time period in order to build sufficient statistical evidence to provide an accurate concentration measurement. This typically takes a few minutes depending on the application.
Does BactoBox differentiate between viable & non-viable bacteria?
Differentiating viable and non-viable bacteria is a technical challenge for any fast bacteria measurement. With traditional plate counts, the bacteria are given time to grow, which helps determine which are viable and culturable, at least at the given conditions. However, due to the slow growth process this is not an option for a fast measurement. Instead, BactoBox uses membrane intactness to assess viability by probing for intact cell structures electrically and omitting non-intact cells from the count. Thus, the unit of BactoBox is “intact cells/ml”. So, is membrane intactness a good estimation of viability? Yes – in the vast majority of cases. Viable cells have intact cell membranes, and most non-viable cells do not have an intact cell membrane – although this can vary depending on how the bacteria have been killed.
Can BactoBox do species differentiation?
No. At this stage in our technology maturation we are not able to differentiate between different species of bacteria. BactoBox measures the total number of bacteria in a sample.
Does BactoBox also measure yeast and molds?
BactoBox measures intact cells/ml. However, BactoBox only measures intact cells in the size range 0.5 µm to 5 µm due to the design of the microfluidic flow cell. This size range covers by far most bacteria and excludes by far most yeast and mold cells. However, in microbiology there will always be edge cases. If a yeast or mold cell is smaller than 5 µm there is a chance it will be counted as an intact cell. We do not recommend using BactoBox to measure yeast and mold cells if yeast and mold counts is the main parameter of interest and if bacteria cannot be used as a proxy. We have vague plans about developing a yeast and mold version of BactoBox in the future and we are still very interested to get input from potential users on where they would want to use such a device.
Is BactoBox an online system or can you make it online?
BactoBox is not an online sensor in the sense it cannot do automatic. BactoBox is a grab sample system that you can use to measure samples from many different sampling points. Regular cleaning of BactoBox is necessary and this feature has not been automated and we are not planning to make a fully online sensor anytime soon. There are many reasons for this, but the primary being that making a fully functional online sensor is much more difficult than what you would think. And believe us, we did try in 2014-2016. However, never say never. The technology is very suitable for being online. It is just not on the roadmap yet.
Is there a consumable?
Yes. The flow cell in BactoBox is a consumable. It lasts for several hundred measurements, but it has to be exchanged eventually. We also provide a range of other consumables that can ease your workflow, but these are all optional.
How does BactoBox correlate with (non-specific) CFU such as standard plate count/aerobic plate count/total plate count?
The answer to this question is unfortunately much more complicated than what we would have preferred it to be. We will start by giving the short explanation for experts in the field, and then we also have a more elaborate explanation for the rest of us.
The short version: BactoBox’ measures intact cells which correlates with CFU, but can be skewed by:
- The presence of cells that do not grow on the selected plates such as stressed or dormant cells, non-culturable cells or cells that prefer other growth conditions – these are counted by BactoBox and not by CFU
- The cells’ tendency to cluster – our data indicates that cell aggregates are separated by fluidic shear forces in our flow cell, which has to be specifically addressed in sample preparation when measuring CFU
The slightly longer version: First, it is important to align on what a colony forming unit (CFU) is. A CFU is a unit used in microbiology to estimate the number of viable bacteria in a sample. You only count the bacteria that decide to grow and multiply when you count CFU. This means that CFU does not always correlate with the true number of viable bacteria in a sample. Two effects contribute to the true number of viable cells being different from the CFU count:
- Bacteria may be viable and still not grow on the plates. This can be caused by bacteria being in a stressed or dormant state. Or the bacteria in the sample can be very selective about when they grow, e.g. by requiring an environment without oxygen, only growing at specific temperatures, requiring specific nutrient compositions or requiring a long time to multiply enough to be visible on plates.
- One colony is not necessarily formed by one bacterium but can be started by multiple bacteria that are clustered together.
In contrast to CFU, the technology in BactoBox counts all bacteria in the sample as long as the bacteria have intact cell membranes. Like any other fast method, BactoBox is not dependent on growth and multiplication of bacteria. This means BactoBox also counts stressed or dormant bacteria cells, or bacteria cells which require exotic growth conditions.
Clustering of bacteria is a well-known problem when measuring CFU as it will typically be several clustered bacteria that result in one colony. Our data indicates that BactoBox break up clusters of bacteria due to the fluidic shear forces in the flow cell. As a result, four bacteria might result in one CFU, but in four intact cells when measured with BactoBox.
Therefore, the correlation between BactoBox and CFU will be linear for samples that:
(1) only contain bacteria in a culturable state, i.e. ready to grow on plates – typically single species samples
(2) cluster minimally
This will often be the case in fermentations and biotech applications where bacteria are grown under controlled conditions.
It is a different story when measuring samples with complex mixes of bacteria in different states and with different preferred growth conditions. These cases include environmental, product or water samples. BactoBox measures the actual cell count in the sample regardless of the mix of bacteria. In contrast, the CFU result strongly depends on the fraction of bacteria which prefer the growth conditions provided to them. To illustrate this point, if you take a sample, split it into two and incubate it on two growth media containing different nutrients, then you will get two different CFU results. Therefore, when measuring on complex bacteria mixes, CFU will usually measure much fewer bacteria than BactoBox – in some cases up to 200x fewer. This makes the correlation between the two methods less apparent, and instead you must consider if measuring the true cell count provides value to your process. However, why not measure everything if you are trying to assess the total bacteria count?
Pitfalls and disadvantages with OD600
Within spectrophotometry, OD600 or nephelometry methods are often used as a time-saving proxy measurement for biomass concentration. The measurement principle is based on light-scattering as shown in the illustration below: Light is passed through the liquid medium and collides with individual bacteria and other particles in the suspension. Higher bacterial concentrations will result in less light reaching the detector.
The wavelength 600 nm is typically chosen because it offers an acceptable tradeoff between signal strength and specificity where most of the light “loss” is caused by light scattering and not by pigment absorption. While OD600 is possible to multiplex in 96-well plates, extremely cheap, simple, and can be performed on-line, there are obvious pitfalls and drawbacks for quantification of microorganisms:
- OD600 provides a number with an arbitrary unit. It does not provide a bacterial concentration unless laborious OD600/CFU calibration curves are carried out.
- Spectrophotometer configurations vary and therefore the OD600 measurements from one device can’t be compared directly with another device unless a calibration with measurement standards is performed |3|.
- OD600 does not differentiate between bacteria and other particles. If the medium contains a background of non-bacterial particles or a high background of dead cells, this could result in misleading results.
- Multiple scattering can lead to non-linear effects where the incident light reaches the detector because it “bounces off” several particles in solution.
- Many bacteria produce pigments that absorb in the OD600 range. The pigment production can vary over time and therefore the OD600/CFU ratio will be inconsistent.
- Bacterial cell size typically vary during a growth curve and because larger cells spread light more than smaller bacteria, this will also give a variable OD600/CFU ratio during the growth curve.
- Some bacteria – especially photosynthetic – transmit light very effectively through the cells and are therefore sometimes referred to as “OD-transparent” because even very high concentration of cells (>109 CFU/mL) result in very little light scattering |2|. This means that they are difficult to detect by OD600 measurements.
Advantages with the BactoBox
While OD600 is based on simultaneous scattering by several bacteria/particles in solution, BactoBox counts individual cells as illustrated below. BactoBox exploits a novel, label-free, impedance flow cytometry technology for cell counting. Objects larger than bacteria (>5 µm) are diverted away from the measuring using microfluidic principles, so they do not lead to clogging or interference with measurements. Bacteria enter the measuring channel, where sets of electrodes detect the individual bacteria on a single-cell basis. Because the detection is based on perturbance of electrical fields, pigmentation or other optical phenomena do not obscure the results and even OD-transparent cells can’t hide from the BactoBox. The electrical signatures for bacteria differ substantially from that of other small micrometer-sized objects and therefore the BactoBox has high specificity for bacteria with intact membranes. These are referred to as intact cells and the BactoBox measurement will report the intact cell concentration (ICC) as a subset of the total particle concentration (Total).

Illustration of differences in the measurement principles. OD600 is based on light scattering by multiple bacteria and other particles. BactoBox measures the fingerprint of the individual intact bacteria in a flow.
Head-to-head comparison of BactoBox and OD600 methods
In theory, these properties should make BactoBox a more reliable proxy method for plate counts for actively growing bacteria. But the proof is in the pudding, and therefore, we set out to investigate this in practice with simultaneous head-to-head measurements on a classical E. coli growth curve:
- Which technique provides the best correlation with plate count during the growth curve?
- Which technique reveals active growth earliest?
BactoBox gives reliable bacterial concentrations in minutes
All measurements were performed in technical triplicates from the primary sample, i.e. the replicates were freshly diluted from the homogenized shake flask sample at the given incubation time.
At first glimpse on the normal axis representation below, both ICC and OD600 tracks growth relatively well. However, if the duration from 3-4 hours is inspected, OD600 somewhat exaggerates the growth relative to CFU and BactoBox measurements. BactoBox ICC is very consistent with the CFU concentration except for the 6.4 hour time point where the large standard deviation on the CFU/mL is the most likely explanation for the offset. On the log10 axis representation, there is excellent correlation between ICC and CFU. On the other hand, at one of the most critical time points (6.4 hours), the high standard deviation on the OD600 curve means that it is not possible to determine if the cell counts are increasing or growth has halted.
Based on these data, BactoBox provides better guidance for the operator at the most critical time points. On the contrary, OD600 has relatively high uncertainty. In addition, BactoBox provides an actual cell concentration and not just an arbitrary unit that needs calibration factors.

Figure 2: BactoBox ICC provides values close to CFU concentrations. Left graph shows the measurements depicted on a normal axis, while the right graph depicts ICC and CFU concentrations on a log axis. All data points are average values of triplicate measurements, where a new dilution series was started for each replicate.
BactoBox detects growth earlier than the OD600 method
One of the most frequent questions when starting a fermentation reactor is “are my cells growing or not”? Several things can go wrong (medium, acid/base control, inoculum, growth-inhibiting substances etc.), and if the cells are not growing you want an answer as quick as possible to avoid wasting further time. In fact, “quick” and sensitive is not enough – you also want robust measurements with a stable baseline as otherwise an increase in the signal could simply be due to “noise”.
In the figure below, the earliest sampling points of the E. coli growth curve are inspected in greater detail in the bar charts below. Both the plate count method and BactoBox measurements show a highly significant (p < 0.01) increase in bacterial concentration for the 40min. sample. On the other hand, with the OD600 measurements, the variation (error bars) is so high that it is not possible to conclude if the cells are growing or not before the 75 min. sampling point. In addition, the numerical increase in OD600 at 75 min. is very small relative to the 10 min. measurement - as an operator, this could likely be interpreted as “no growth yet”.
Another classical pitfall with OD600 is that the (supposedly sterile) medium used to “zero” the spectrophotometer can be contaminated by growth. This could make initial growth seem to have even lower OD600 values than the blank.
In brief, the BactoBox is significantly more sensitive at detecting early growth at low bacterial concentrations than the OD600 method.

Figure 3: BactoBox reflects an increase in growth earlier than OD600: Comparison of measurement principles for the earliest sampling points of the E. coli growth curve. Statistical analysis shows a highly significant increase already at the 40 min point for ICC and CFU concentrations (Student’s T-test, p < 0.01). For OD600 the cell density has not increased significantly before the 75 min. sampling point (p < 0.05).
BactoBox correlation with CFU is consistent over time
Because BactoBox detects single events as they pass by the electrodes in the measurement channel, it is not affected by the shortcomings of many optical cuvette-based light-scattering methods. This is indeed reflected in the figure below where the BactoBox ICC is very consistent over time with almost a perfect 1:1 correlation with plate counts. On the other hand, OD600 to CFU ratio is not consistent over time which is clearly shown with the red curve where the calibration factor changes from 0.8*109 to 6.4*109 over time, i.e., a factor 8 difference during the bacterial growth curve.
The offsets in the OD600/CFU ratio over time can be a problem because OD600 is very often used in conjunction with CFUs to create a calibration curve for test suspensions in e.g. antimicrobial studies. Once the calibration curve has been established the analyst will use this correlation as a proxy to determine the here-and-now concentration of CFU/mL before performing experiments. The data in the present investigation show that there are serious pitfalls with this approach. For example, a calibration curve for an 11-hour (overnight) culture is not valid for a 4-hour culture since the CFU/mL concentration would be overestimated by 200%.
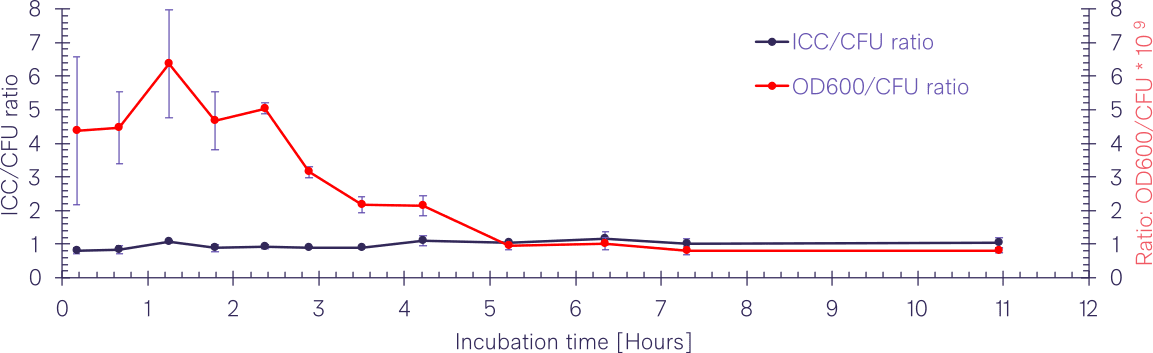
BactoBox has an almost perfect 1:1 correlation to plate counts. The intact cell concentration correlates closely with CFU concentrations. This is not the case for OD600.
BactoBox as a superior method for in-process bacterial samples
Here we provided a head-to-head comparison of BactoBox intact cell concentrations and OD600 for tracking the growth of a shake flask culture of E. coli. This comparison clearly showed that BactoBox is a fast and reliable growth tracker for in-process samples of actively growing bacteria.
- BactoBox is more sensitive than the OD600 method. In practice this means that BactoBox can detect growth much sooner than OD600.
- The accuracy (similarity to the plate counts) of the BactoBox method is better than that of the OD600.
- The correlation of BactoBox intact cell concentration relative to plate counts is consistent over the entire incubation period.
- BactoBox provides absolute concentrations for bacteria, not arbitrary units that must be converted via calibration curves.
References
- USP <1223>: Validation of Alternative Microbiological Methods. United States Pharmacop. 04-Oct-2021, 4–6 (2021).
- Myers, J. A., Curtis, B. S. & Curtis, W. R. Improving accuracy of cell and chromophore concentration measurements using optical density. BMC Biophys. 2013 61 6, 1–16 (2013).
- Stevenson, K., McVey, A. F., Clark, I. B. N., Swain, P. S. & Pilizota, T. General calibration of microbial growth in microplate readers. Sci. Reports 2016 61 6, 1–7 (2016).
Enumerate Bacteria with BactoBox®
Enumerate Bacteria with BactoBox®
Enumerate Bacteria with BactoBox®
Please submit your contact and application details using the form below. A specialist from our team will contact you within one business day to discuss the technical compatibility of BactoBox® with your needs and outline potential trial opportunities. Alternatively, reach out directly to our CEO, Gustav.
Learn More or Request a Trial
Learn More or Request a Trial
Learn More or Request a Trial
Copyright © 2023 SBT Instruments A/S. All rights reserved.
